Overview
The WILDLABS Virtual Meetup Series is a program of webinars for community members and wider partners to discuss emerging topics in conservation technology and leverage existing community groups for virtual exchange. The aim of the series is to bring leading engineers in the tech sector together with academics and conservation practitioners to share information, identify obstacles, and discuss how to best move forward.
Season One of the series took place in late 2018, covering new data collection techniques through Networked Sensors for Security and Human-Wildlife Conflict (HWC) Prevention and Next-Generation Wildlife Tracking, and effective utilization of that information through Big Data in Conservation. Season Two ran during the first half of 2019 and focused on Tools and Spaces for Collaboration, the Low-Cost Open-Source Solutions these approaches are producing, and how to put the information they’re generating to use through Creative Approaches to Data-Driven Storytelling.
Season Three is taking place throughout the second half of 2019 and is exploring the theme of noninvasive monitoring technologies in conservation. This season's topics include Camera Trapping, Drones, Environmental DNA (eDNA), and Acoustic Monitoring. After a more approach-driven second season, we’re eager to be diving back into the realm of development and implementation in the context of these ever-evolving tools.
We are always looking to tailor these meetups to community interests and needs, so if you have ideas about specific discussion points you'd like to see covered during this season please join the thread and share your thoughts.
Meetup 3: Environmental DNA (eDNA)
Date & Time
Thursday, December 5th, 2019
4:00pm-5:30pm GMT / 11:00am-12:30pm EST
Background & Need
Environmental DNA, or eDNA, is a molecular sampling technology that collects information about organisms using DNA shed by them into their environment. While eDNA holds exciting potential to become an efficient, low-cost, non-invasive ecological monitoring method, a lack of standardization of approaches and purpose-built sampling equipment make it hard to compare results and determine its actual effectiveness across studies. Detection success and accuracy also vary greatly among species and environments, making standardization a challenge.
However, as eDNA moves toward becoming an industry standard method for species detection and management, the equipment is beginning to transition from largely do-it-yourself experimental contraptions to professionally engineered tools. For example, Smith-Root Inc. recently developed the first-ever purpose-built eDNA sampling system, which they call ANDe™. We’re also seeing an expansion of eDNA applications from mainly marine to freshwater and even terrestrial environments, although the latter still requires substantial growth.
As this nascent field develops, it is critical for the conservation tech community to explore and identify how eDNA applies to management needs, and to ensure that continued development meets those needs. Beyond indicating species presence, research so far suggests that eDNA can contribute to conservation by deepening understanding of population dynamics, resource usage, disease presence, invasion pathways of non-native species, and population genetics, to name a few. This indicates that it could be a critical tool for managing imperiled and invasive species. This meetup will explore some of these current and future applications of eDNA, articulate how it applies to conservation needs, and define what challenges we face in maximizing its potential.
Outcomes
The aims of this discussion are as follows: to introduce eDNA in conservation; to describe how it is being used, including what needs it is addressing and how it fits into the wider conservation tech ecosystem; to discuss the future of this tool, and to identify the obstacles in advancing its capacity.
Recording
Click through here to watch the full meetup (note: audio transcripts now available with recordings!)
Virtual Meetup Notes
During the third event in season three of our Virtual Meetup Series, more than 65 attendees joined us from at least 18 countries around the world. Thank you to all who came and participated. For those of you who were unable to join live, we’ve recorded the session so that you may view it at your convenience. You can also check out the presentation notes and further reading suggestions below.
Speaker: Kat Bruce
Background
- Tropical ecologist by training, used high throughput sequencing and metabarcoding to generate large datasets to inform environmental management decisions
- Even in 2012-13, literature showed huge promise for these tools, but early papers were published in molecular journals and not shared effectively with environmental managers
- NatureMetrics exists to bridge this gap between research and conservation practitioners
Basics of eDNA
- Barcoding: Sequence a specimen’s DNA for a particular gene and use that sequence to identify the species (1 specimen, 1 sequence); prone to error if other DNA is included in sample
- Metabarcoding: Uses high throughput DNA sequencing, which allows you to take a complex sample containing DNA of many different specimens and generate separate sequences for each. Can also sequence many samples in parallel.
- For the past decade we’ve been sampling water, which contains genetic material shed by animals as they move through it – we can filter out DNA to sequence
- DNA in water breaks down after a few days, so detections give contemporary rather than historic signals
- Sample collection – NatureMetrics has developed a user-friendly kit that enables anyone, anywhere, to collect high quality samples
Processing & Interpretation
- Processing steps:
- Post filter to the lab
- DNA extracted from filter
- DNA of target group amplified
- Sequence on high-throughput sequencer
- Computational process that takes data files of 30 million sequences and summarizes them into a species-by-sample table
- Run sequences against a reference database to identify species
- Interpretation
- Presence/absence is the most certain information, but for some groups like fish that are in the water and shedding DNA at relatively consistent rates the number of sequences we now know that it does tend to correlate with relative abundance
Effectiveness
- Freshwater bioblitz example:
- Consistency of relative abundance data
- Single eDNA sample detects more species than a single electrofishing sample
- Across about 30 studies now we’ve found that one eDNA sample provides better data than many years of electrofishing data combined
- Overnight netting vs 3L eDNA sample in Sweden
Standardization
- Comparison tests this past summer at a conference on eDNA for the Fisheries Society with three highly experienced labs
- Trial: Had everyone bring their normal equipment, take a sample from a pool of local river water they brought in, and process it how they normally would
- Results: all followed same general principles but made different choices at every stage of processing
- Very comparable data for the most abundant species in terms of detection and relative abundance
- With rarer species, we saw more stochasticity and drop out
- Comparing this data with traditional fisheries data from the same river, we found that every species of fish recorded by electrofishing was identified by all three labs.
- Drafting European standard for water sampling for eDNA
- Effort to standardize without being overly prescriptive at this stage
Case Study: eDNA survey with WWF in the Peruvian Amazon
- Wanted to know about river dolphins, Amazon manatees, and a few species of catfish
- Took samples at 40 points across northern Peruvian Amazon 10-15km apart and four replicate samples at each point
- Found higher number of dolphin sequences moving downstream – is it because of DNA accumulation downstream, or because populations are actually greater here?
- Answered this question by looking for same pattern across all species, and found that it was not consistent, meaning that it is likely reflective of real population differences
- With similar effort to surveying just one species, we generated a dataset of over 700 vertebrate species, including many terrestrial mammals that are difficult to monitor with other methods (assisted in this region by frequent rainfall that washes DNA into the river)
Takeaways
- Limitations
- Not possible to count individuals
- No age or size data
- Incomplete reference databases (though improving with time and effort)
- Spatial uncertainty
- Advantages
- Unprecedented amounts of data
- Easy sampling – reduced cost
- Detct hard-to-observe species
- Archive and share samples
- Final thoughts
- Aquatic eDNA is a well-validated and powerful survey method for fish
- It has limitations like any other survey method
- Anyone can collect high-quality samples
Speaker: Alice Valentini
Background
- Research engineer and Co-Founder at SpyGen; focus on aquatic biodiversity monitoring with eDNA
- SpyGen: Founded in 2011, mission-driven company with goal to improve monitoring and conservation of biodiversity through the study of eDNA
- Focus on analysis as well as sample collection kits
Barcoding
- Target specific species with primer
- Study on the Great Crested Newt (Triturus cristatus)
- Compared 4 methods to determine the likelihood of detection
- Results
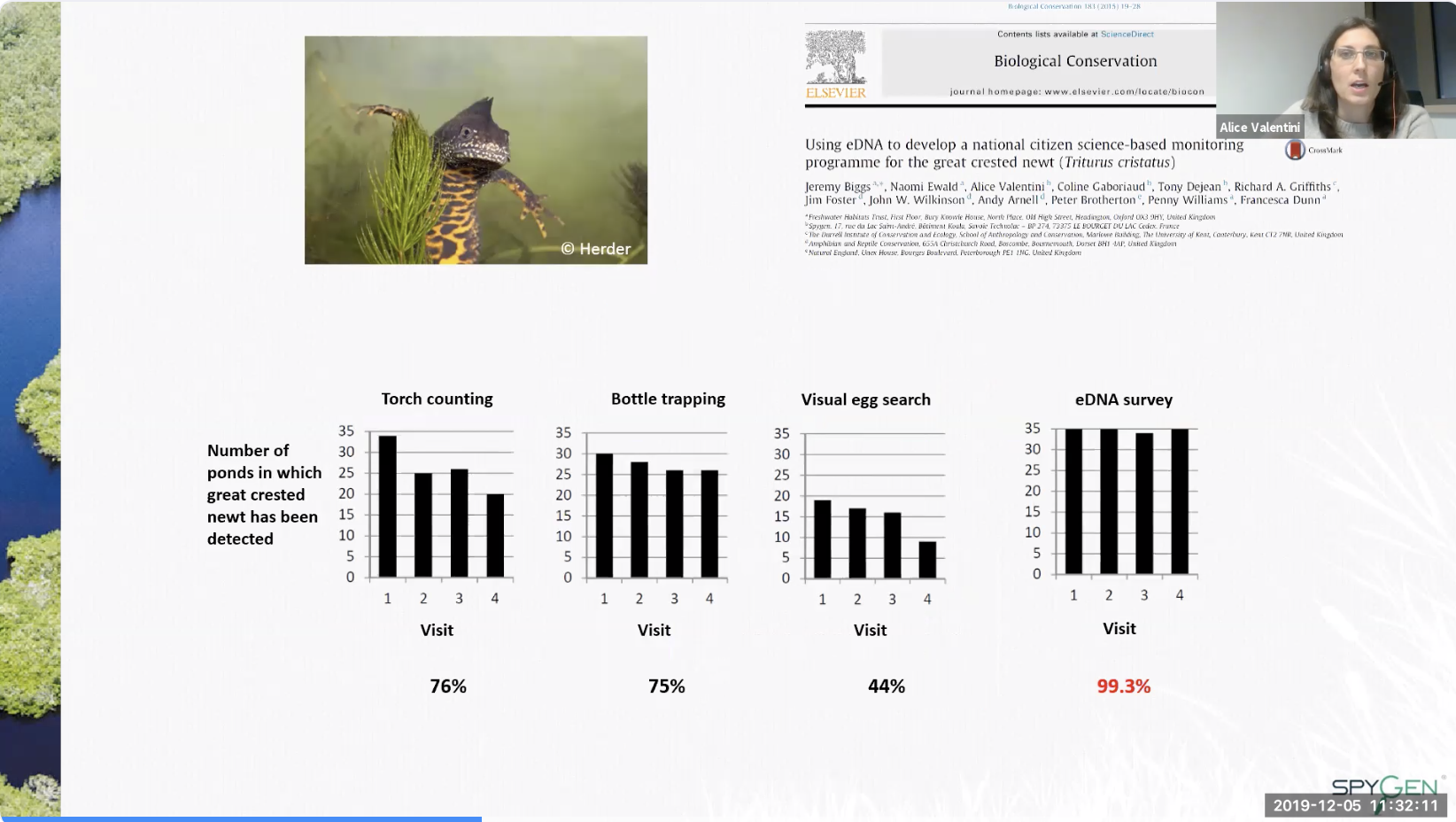
- Citizen science: volunteers not trained or supervised sampled 239 sites and correctly identified 91.3% of sites as supporting newts (meaning techniques are easily implemented in the field and can employ citizen scientists)
Metabarcoding
- Use group rather than species-specific primers, compare data with genetic reference database to determine taxa
- In an assessment of fish diversity, they found that two eDNA samples were equivalent to ten years of electrofishing data
- Also detected 2 critically endangered species, one of which was thought to be locally extinct
Biodiversity Assessments
- French Guiana study
- Sampled river drainage for two rivers roughly every 10k and analyzed for fish and also mammals
- Duplicated French Guiana study in Senegal, where they assessed all vertebrates – here they were able to distinguish sharks, mammals, fish, reptiles, and bird
- Brazilian study using eDNA metabarcoding to describe tropical forest biodiversity
- Described local amphibian populations
- Detected all 9 species known to be present in the region, compared with traditional methods, which normally only detect 3
Detecting Rare and Elusive Species
- Study with WWF in the Mekong river detecting the Mekong Giant Catfish
- Also found the Irrawaddy dolphin
- Detecting the Giant Muntjac in the mountains of Laos
- Also detected the red-shanked douc, Asian black bear, Asian small-clawed otter, and four-eyed turtles, among other species
Conclusions
- eDNA is a powerful tool for species detection
- It gives us the capacity to monitor rare, endangered, or cryptic species
- Also enables early detection of alien species
- eDNA inventories can serve as new benchmarks for monitoring ecosystem health, functioning, and services, and provide strong indicators for early detection of species decline and biodiversity erosion
Speakers: Arnaud Lyet & Robin Naidoo
Limitations of classical methods
- Direct observations only possible with abundant species
- Indirect observations only possible when signs are abundant
- Requires skilled and trained observers
- Even with trained observers, misidentification of signs is frequent
- Surveys of rare/elusive species can be expensive due to technology required (aerial surveys, GPS collars, camera traps) and field effort
- Individual ID/ monitoring often challenging, sometimes impossible
What can eDNA do?
- Aquatic and terrestrial biodiversity inventories
- Terrestrial biodiversity from what source?
- Identify species from its track?
- Identify a particular individual from its track?
- Cost-effectiveness?
Individual Identification
- Polar bears
- Traditional methods require sedation and physical sample collection
- Idea: Use Next Generation Sequencing to be able to identify an individual based on eDNA collected from their pawprint
- Results: Still needs work, only the artificial pawprint (from a deceased bear) resulted in full genotyping, but mitochondrial DNA collected from wild samples
- Sumatran rhino
- Swabbed mud from footprints of rhinos
- Mitochondrial DNA successfully extracted from 30% of samples
- Confirmation of species from footprint to avoid confusion with other species like tapir
- Next steps: Explore possibility to retrieve nuclear DNA for genotyping
Global Biodiversity Inventory
- More complicated for terrestrial than for aquatic? How well does it work for very rare species?
- Sumatran rhino in West and Central Kalimantan
- Preliminary tests from water samples in an area known to contain rhino detected many aquatic and terrestrial species, but no rhino
- Opportunities to monitor tigers and their prey?
- Sumatran rhino in West and Central Kalimantan
- The problem:
- Most eDNA studies focus on detection of species, rather than relative abundance
- Few studies have compared eDNA abundance estimates with other methods, and these have only focused on aquatic species
- eDNA sampling issues for terrestrial vertebrates remain largely unresolved
Comparing wildlife abundance from camera traps and eDNA
- Camera traps are a standardized, well-understood way to measure relative abundance and sometimes densities of wildlife species – good for comparison
- Questions:
- What is the relationship between abundance estimates from camera traps and eDNA sampling?
- How does the quantity of eDNA detected in the water relate to the relative abundance of the species in the watershed?
- Area: South Chilcotin Mountains in western Canada, half within a park and half outside of it; variety of streams and rivers to collect eDNA samples (in two separate watersheds); fairly large, in-tact wildlife assemblage but with human presence throughout it
- Methods:
- 61 camera traps set up in a grid covering 800km2 and spaced at ~3km; ran from mid-May – mid-September of 2018
- eDNA samples collected with Vampire Sampler; filtered river water for 30 min periods across entire area covered by camera traps
- eDNA abundance for a variety of species compared across watersheds and to number of camera trap detections
- Results:
- Distribution of detections across taxa varied between camera traps and eDNA
- eDNA was best at detecting small mammals, and mule deer, while camera traps did better at detecting cat species
- There was a relationship between the number of copies of DNA in samples and the number of pictures from camera traps, but need more replicates to better understand it

- Distribution of detections across taxa varied between camera traps and eDNA
- Next steps:
- Now collecting more data at same locations to see how the relationship changes over time
- Eventually plan to use similar methodology to study tigers and their prey in SE Asia
The promise of eDNA
- Potential for non-invasive sampling of biodiversity
- Novel technology still in development but promises to revolutionize our understanding of biodiversity (locations and abundance)
- Techniques and costs likely to improve in coming years due to rapid technological advances
Further Reading
Other links referenced in the live chat:
- Footprint Identification Technique (FIT), another non-invasive monitoring technique developed by from WildTrack, and their project ConservationFIT.
- This paper by Maestri et al. highlighting the possibilities of MinION.
- An article from 2018 announcing GBIF's intentions to integrate eDNA data, and this update from 2019.
- Srivasthan et al. paper on using MinION sequencing for large-scale species discovery (they sequenced 7,000 species), and an older paper from the same authors on where they sequenced 500 samples on one flow cell.
Next Steps
- Jump over to this thread to continue the conversation or ask follow-up questions from the event.
- Look out for information regarding our next meetup on acoustic monitoring in January of 2020!




Add the first post in this thread.